Karoline Heiwolt1, Cengiz Öztireli2, Grzegorz Cielniak1
1University of Lincoln 2University of Cambridge
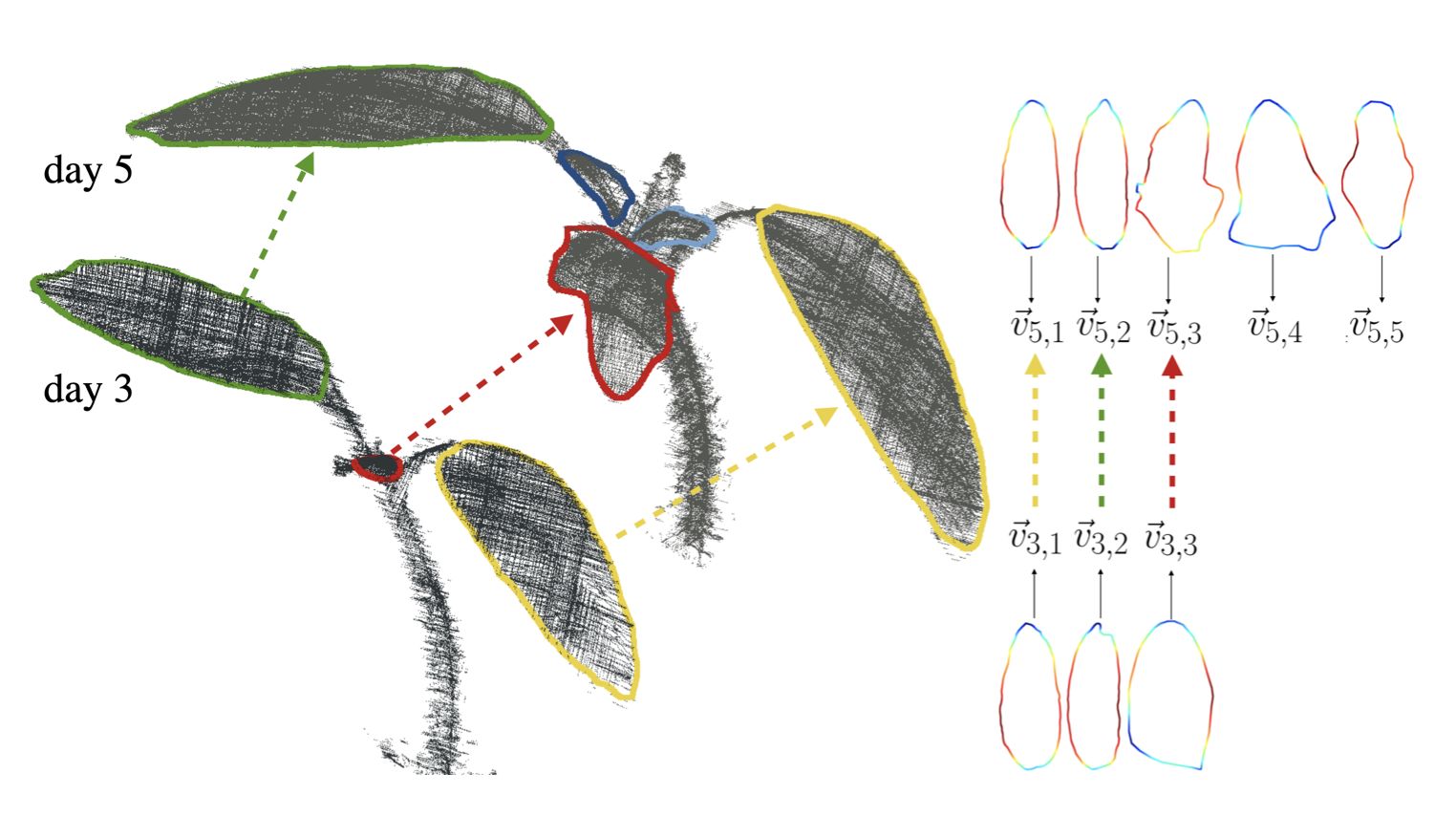
Plants are dynamic organisms. Understanding temporal variations in vegetation is an essential problem for all robots in the wild. However, associating repeated 3D scans of plants across time is challenging. A key step in this process is re-identifying and tracking the same individual plant components over time. Previously, this has been achieved by comparing their global spatial or topological location. In this work, we demonstrate how using shape features improves temporal organ matching. We present a landmark-free shape compression algorithm, which allows for the extraction of 3D shape features of leaves, characterises leaf shape and curvature efficiently in few parameters, and makes the association of individual leaves in feature space possible. The approach combines 3D contour extraction and further compression using Principal Component Analysis (PCA) to produce a shape space encoding, which is entirely learned from data and retains information about edge contours and 3D curvature. Our evaluation on temporal scan sequences of tomato plants shows, that incorporating shape features improves temporal leaf-matching. A combination of shape, location, and rotation information proves most informative for recognition of leaves over time and yields a true positive rate of 75%, a 15% improvement on sate-of-the-art methods. This is essential for robotic crop monitoring, which enables whole-of-lifecycle phenotyping.
Links:
Paper